Plumage phenotypes, mitochondrial DNA and nuclear DNA tell different stories
LINKED PAPER
Discordance between genomic divergence and phenotypic variation in a rapidly evolving avian genus (Motacilla). Harris, R.B., Alström, P., Ödeen, A. & Leache, A.D. 2018. Molecular Phylogenetics and Evolution. DOI: 10.1016/j.ympev.2017.11.020. VIEW
Wagtails (genus Motacilla) are widespread across the Old World. Most of the species show pronounced geographical plumage variation, especially the Yellow Wagtail Motacilla flava and White Wagtail M. alba complexes, which are renowned for their many different-looking subspecies (Alström & Mild 2003; Fig. 1).
Previous studies based on mitochondrial DNA (mtDNA) have suggested unexpected phylogenetic relationships and phylogeographic groups that are inconsistent with traditional subspecies (defined by plumage), as well as generally no or very slight genetic differentiation among traditional subspecies (Alström & Ödeen 2002, Ödeen & Björlund 2003, Pavlova et al. 2003, 2005, Alström et al. 2015, Li et al. 2016, Drovetski et al. 2018).
Complete species-level phylogeny
In our recently published paper, we investigated the link between phenotypes and genotypes in a phylogenetic context by using genome-wide single nucleotide polymorphism (SNPs), nuclear introns, and mtDNA. Unlike previous studies, we included all species and most of the subspecies in the genus, with emphasis on the three phenotypically most diverse Palearctic species (White, Yellow and Citrine M. citreola).
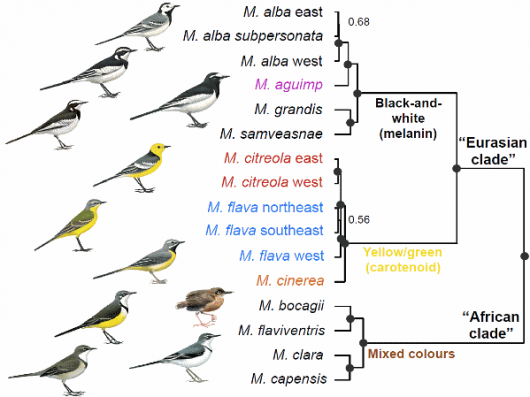
SNPs provide a robust estimate of species-level relationships (Fig. 1). The SNP phylogeny is mostly corroborated by a time-calibrated species tree based on mtDNA and nuclear introns (Fig. 2). We jointly consider these trees in our interpretation of wagtail relationships – while the SNP tree lacks data from the White-browed Wagtail M. maderaspatensis, it provides sampling from across the entire genome from many more individuals than the mtDNA-intron tree.

The trees identify two primary clades, one comprising all of the Eurasian species plus the African Pied Wagtail M. aguimp and one containing the Afrotropical and Madagascar species. The former clade is further subdivided into one clade containing the species with black-and-white, melanin-based, plumages and one including the species with yellow/green, carotenoid-based, plumage colours (Fig. 1).
A previous study had suggested that a species traditionally classified as a sylvioid, the São Tomé Shorttail Amaurocichla bocagii, is a forest-dwelling wagtail (Alström et al. 2015). Our study confirms that it is indeed a wagtail, despite that it differs dramatically from its congeners in appearance, habitat choice and behaviour (Alström et al. 2015), and should therefore be placed in the genus Motacilla. Being endemic to São Tomé off west Africa, it may seem surprising that it is most closely related to the Madagascar Wagtail M. flaviventris.
Mitochondrial DNA tree is incongruent with other data
The mtDNA-only (Fig. 3) tree is strongly incongruent with both the SNP tree and the mtDNA-intron species tree Furthermore, it is in conflict with morphological evidence. In particular, M. aguimp is placed outside the ‘black-and-white clade’, and there are three separate clades that include different geographical populations of both M. flava and M. citreola (indicated by X, Y, Z in Fig. 3). We conclude that the mtDNA tree misrepresents the species phylogeny, and has probably been affected by past hybridisation and introgression of mtDNA.

Recent, rapid divergence of Palearctic taxa
According to our time estimate, the extant wagtail lineages originated during the Pliocene. The ‘African clade’ started to diverge around the Pliocene/Pleistocene border, while the Eurasian clade underwent rapid diversification during the latter half of the Pleistocene. This is considerably younger than previous, less complete, studies which estimated the split between the primary clades at 4.5–5 mya (Voelker 2002, Alström et al. 2015). Potential differences between these discrepancies are discussed.
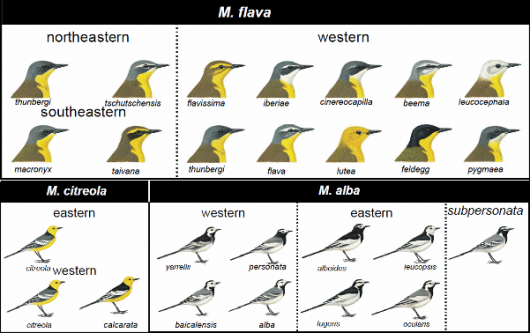
The rapid divergence, perhaps in combination with ongoing or past gene flow, is reflected by the lack of genetic differentiation between phenotypically distinct subspecies within the M. alba, M. flava and M. citreola complexes. Instead, the SNP data support geographical regions, each of which is home to two or more different looking subspecies (Fig. 4). Both the M. flava and M. citreola complexes exhibit an east-west divide that contradicts both subspecies taxonomy and phenotypic variation (Fig. 4).

Apparently, the plumage divergence has been extremely fast and has resulted in several cases of parallel evolution. For example, the ‘thunbergi phenotype’ (thunbergi, macronyx) is found in all three M. flava populations, while the ‘flava phenotype’ is found in both the western (flava, beema) and northeastern (tschutschensis) populations. Furthermore, taxa with bright yellow supercilium are found in both western (M. f. flavissima, M. f. lutea) and southeastern (M. f. taivana) clades, including in widely disjunct geographical areas within the western clade. Motacilla aguimp and Mekong Wagtail M. samveasnae are remarkably similar-looking despite being geographically and genetically far separated. It seems likely that the colour/pattern differences among taxa can be switched on and off in different combinations through a rather simple system. Future research will hopefully shed light on this system. The extremely odd appearance of M. bocagii has previously been suggested by Alström et al. (2015) to be due to adaptation to a novel habitat.
Taxonomic implications
Our SNP data support a monophyletic M. flava complex consisting of three clades with barriers to gene flow (in agreement with mtDNA; Pavlova et al. 2003), whereas combined analysis of nuclear introns and mtDNA recover M. flava as paraphyletic. While it is likely that our SNP data reflect the true species relationships, it is clear that these two species have shared a complex and recent history. At any rate, M. citreola is recovered as monophyletic in all analyses.
The rare endemic Moroccan White Wagtail subspecies subpersonata stands out as distinct in both plumage, mtDNA, and SNPs, and therefore warrants further study.
References and further reading
Alström, P. & Mild, K. 2003. Pipits and Wagtails of Europe, Asia and North America. A&C Black, Princeton University Press.
Alström, P. & Ödeen, A. 2002. Incongruence between mitochondrial DNA, nuclear DNA and non-molecular data in the avian genus Motacilla: implications for estimates of species phylogenies. In: Alström, P. (Ed.), Species Limits and Systematics in Some Passerine Birds. Uppsala University [Ph.D. thesis].
Alström, P., Jønsson, K.A., Fjeldså, J., Ödeen, A., Ericson, P.G.P. & Irestedt, M. 2015. Dramatic niche shifts and morphological change in two insular bird species. Roy. Soc. Open Sci. 2: 140364. VIEW
Drovetski, S.V., Reeves, A.B., Red’kin, Y.A., Fadeev, I.V., Koblik, E.A., Sotnikov, V.N., & Voelker, G. 2018. Multi-locus reassessment of a striking discord between mtDNA gene trees and taxonomy across two congeneric species complexes. Molecular Phylogenetics and Evolution 120: 43–52. VIEW
Li, X., Dong, F., Lei, F., Alström, P., Zhang, R., Ödeen, A., Fjeldså, J., Ericson, P.G.P., Zou, F. & Yang, X. 2016. Shaped by uneven Pleistocene climate: mitochondrial phylogeographic pattern and population history of white wagtail Motacilla alba (Aves: Passeriformes). J. Avian Biol. 47: 263–274. VIEW
Ödeen, A. & Björklund, M. 2003. Dynamics in the evolution of sexual traits: losses and gains, radiation and convergence in yellow wagtails (Motacilla flava). Mol. Ecol. 12: 2113–2130. VIEW
Pavlova, A., Zink, R.M., Drovetski, S.V., Red’kin, Y. & Rohwer, S. 2003. Phylogeographic patterns in Motacilla flava and Motacilla citreola: species limits and population history. Auk 120: 744–758. VIEW
Pavlova, A., Zink, R.M., Rohwer, S., Koblik, E.A., Red’kin, Y.A., Fadeev, I.V. & Nesterov, E.V. 2005. Mitochondrial DNA and plumage evolution in the white wagtail Motacilla alba. J. Avian Biol. 36: 322–336. VIEW
Semenov, G.A., Scordato, E.S.C., Khaydarov, D.R., Smith, C.C.R., Kane, N.C. & Safran, R.J. 2017. Effects of assortative mate choice on the genomic and morphological structure of a hybrid zone between two bird subspecies. Mol. Ecol. 26: 6430–6444. VIEW
Semenov, G.A., Koblik, E.A., Red’kin, Y.A., & Badyaev, A.V. 2018. Extensive phenotypic diversification coexists with little genetic divergence and a lack of population structure in the White Wagtail subspecies complex (Motacilla alba). Journal of Evolutionary Biology in press. DOI: 10.1111/jeb.13305. VIEW
Image credits
Featured image: Citrine Wagtail Motacilla citreola citreola Xinjiang, China, May 2018 © Per Alström
Figure 1: Top: Species in “Eurasian clade” © Bill Zetterström from Alström & Mild (2003); M. bocagii © Jon Fjeldså; other species in “African clade” Ren Hathway © Lynx Edicions (from del Hoyo et al. 2004. Handbook of the Birds of the World, Vol. 9).
Bottom: © Bill Zetterström from Alström & Mild (2003)
Blog posts express the views of the individual author(s) and not those of the BOU.
If you want to write about your research in #theBOUblog, then please see here.